NanoString GeoMx Spatial Transcriptomics
The recommended and preferred solution for best-in-class GeoMx data analysis
NanoString nCounter Gene and miRNA Expression
Bruker's recommended and preferred solution for best-in-class nCounter data analysis
Bulk RNA-seq Gene Expression
DIY Bioinformatics for this genomics staple

CS Genetics SimpleCell 3' Gene Expression
Single Cell Without the Hassle (or Instrument)
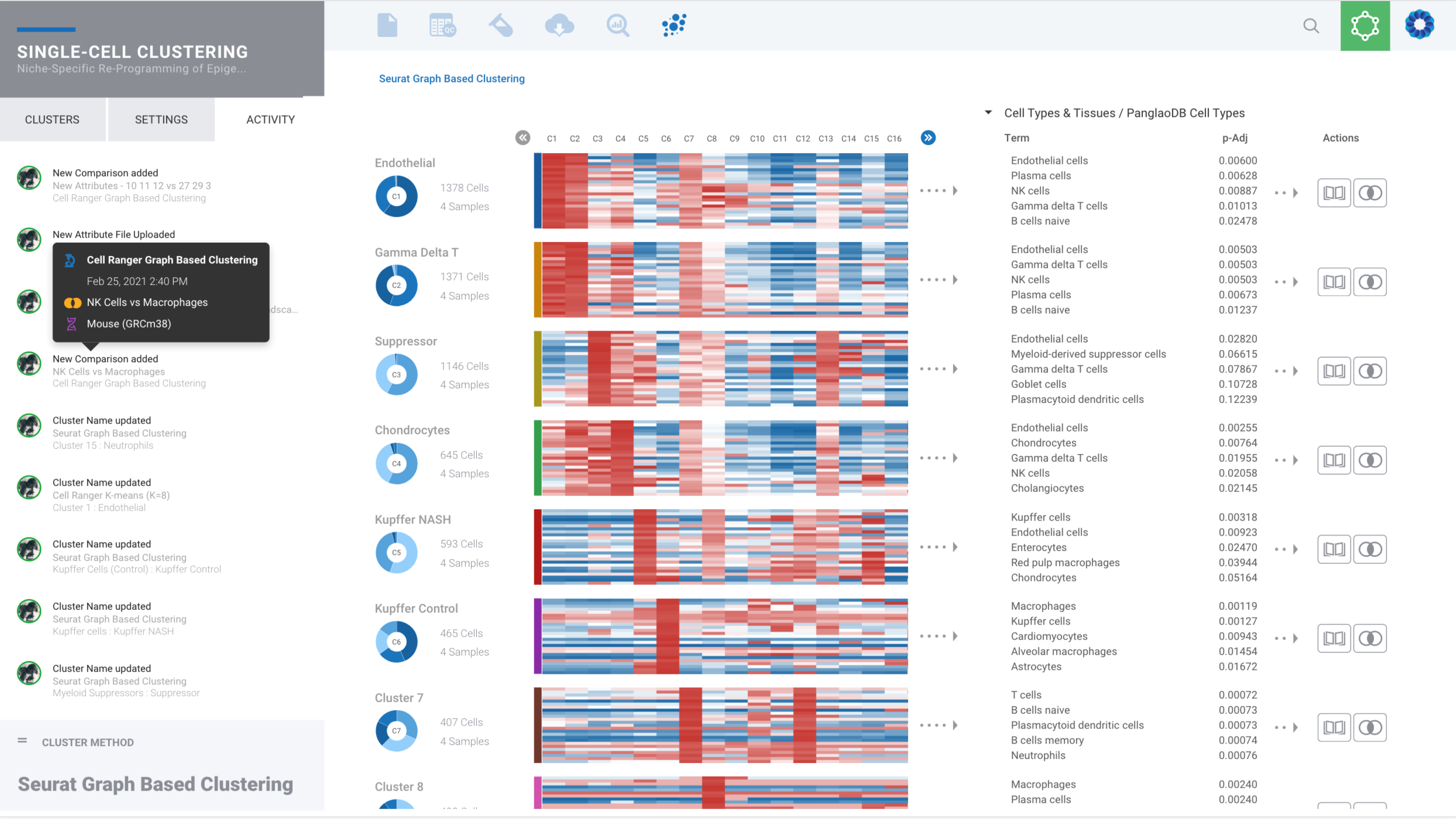
Single Cell Gene Expression
From FASTQ to cell clusters and beyond
Gene Regulation & Anti-Sense: Small RNA-seq
Small RNA-seq data analysis designed for the biologist
Histone Mark & Transcription Factor: ChIP-seq
Comprehensive ChIP-seq data analysis

Chromatin Accessibility: ATAC-Seq
Genome-wide chromatin accessibility analysis

Sharing & Collaboration
Accelerate teamwork anywhere in the world

Knowledge Graph and Search
Data organized for semantic queries

COVID-19 Diagnostic Monitoring System
SARS-CoV-2 Viral Mutation Tracking
COVID Research Community
Global research analyzing COVID-19 genomic datasets
Immuno Oncology
Making discoveries on IO research
Breast Cancer
Identifying differentially expressed miRNAs in breast cancer
HIV
Transcriptional and genomic profiling study of HIV+ DLBCL
Diabetes
Discover and accelerate T1D
Multi-Omics Dataset
Explore multi-omics datasets in epigenetics research

Single Cell Data
Explore datasets with our Cell Ranger graph-based clustering

nanoString Gene Expression
Analyze your nCounter data in minutes

nanoString Mouse Glial Profiling Panel
Explore neuroinflammatory astrocyte subtypes in the mouse brain

RNA-Seq Data
Get started with bulk RNA-Seq data
Subscriptions and Pricing
Learn about Enterprise, Professional, and Academic subscriptions
Enterprise Offering
Expand the depth and reach of your scientific data teams

 Learn More
Learn More