The ROSALIND Tracker is a real-time dashboard for the United States rapid genotyping "Project ROSA" program for SARS-CoV-2. This research was supported by the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health as part of the Rapid Acceleration of Diagnostics (RADxˢᵐ) initiative.
SAN DIEGO, CA – June 29, 2022 – ROSALIND®, a software leader in extracting meaningful insights from diverse pools of life science data, today announced the launch of a web-based platform, The ROSALIND Tracker for monitoring the emergence of SARS-CoV-2 variants, including Delta, and the latest variant of concern, Omicron. Through collaborative efforts from Thermo Fisher Scientific, Helix, Aeigis, and The University of Washington Medicine, The ROSALIND Tracker was developed as part of the rapid genotyping "Project ROSA" program for SARS-CoV-2 and has been featured in a recent Press Release with Thermo Fisher and one by the National Institutes of Health published today.
Since the emergence of SARS-CoV-2 in early 2020, the virus has mutated to generate variants that can be more transmissible, replicate faster resulting in increased viral loads, and potentially evade vaccine neutralization. It is critical to monitor the genetic lineages within positive SARS-CoV-2 samples so that there is information about the landscape of variants circulating within a region and the ability to quickly identify variants that have the potential to evade vaccines or cause more severe diseases.
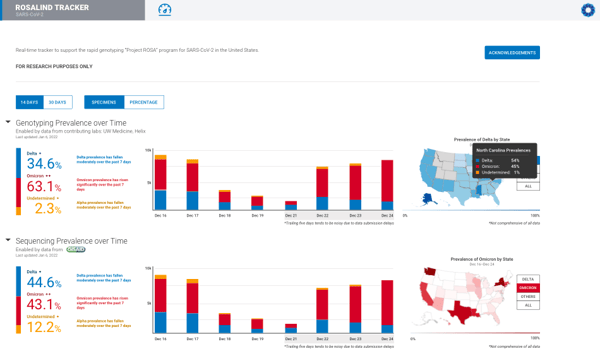
The ROSALIND Tracker allows users to track real-time updates of SARS-CoV-2 outbreaks and identifies the different variants of concern, such as Delta or Omicron. Users will also be able to track genotyping and sequencing prevalence over time. The public can access The ROSALIND Tracker at no cost.
Additionally, as part of "Project ROSA's" global research efforts, the team has collaborated on a publication focused on methods for SARS-CoV-2 detection and the monitoring of circulating variants, such as Delta and Omicron. The research paper, published in the Journal of Clinical Microbiology: A method for variant-agnostic detection of SARS-CoV-2, rapid monitoring of circulating variants, detection of mutations of biological significance and early detection of novel and emergent variants such as Omicron, utilizes real-world data from GISAID, including genome sequences and metadata, which were used for the selection of markers to identify the different COVID-19 variants, while the data analysis for identifying SARS-CoV-2 markers was performed using ROSALIND'S Variant Analysis for Diagnostic Monitoring (DxM).
The study aimed to identify SARS-CoV-2 markers useful for the detection of positive COVID-19 samples across all major WHO identified variants, to develop a panel of SNP markers that can be used to accurately assign lineages to COVID-19 positive samples, and to implement a genotyping approach for the early detection of new and re-emerging variants that signals when updated markers for rapidly emerging variants need to be added. Findings from the publication can be found here.
This research was supported by the National Institute of Biomedical Imaging and Bioengineering or the National Institutes of Health as part of the Rapid Acceleration of Diagnostics (RADxˢᵐ) initiative. We gratefully acknowledge the data contributors and patients offering sample data to the GISAID Initiative. Specific acknowledgment for data curation goes to the Authors, the Originating Laboratories obtaining the specimens, and the Submitting Laboratories that generated genetic sequences and metadata. We would also like to note our appreciation for the EpiCov Data Curation Team within GISAID mentioned here.
About ROSALIND
Based in San Diego, ROSALIND enables the world’s life science researchers to derive more value from their data and complete projects faster. We believe the potential of biomedical and multi-omic data is maximized when it is accessible, interactive, and shareable. Extracting meaningful insights from pools of data takes a truly interoperable ecosystem that breaks down silos of complex information and sparks collaboration. The ROSALIND platform harnesses the power of scientific collaboration, cloud computing, and data science to enable integrated analyses that unify knowledge through an intuitive user interface that is built for any researcher. For more information, please visit www.rosalind.bio.










.png)
