Could health outcomes be determined by the little guys who call us home?
The San Diego MicroBiome Meetup, hosted by Clotide Telling of Illumina, Inc., kicked off with a presentation by OnRamp's Scientific Advisor, Rob Edwards. Rob's story of the discovery of the crAssphage is a truly amazing example of Metagenomics and the MicroBiome, and is testament to the power of the sequencing revolution.
By now we’ve all heard the claim that each of us hosts more bacterial cells than our own cells. Though this extreme situation may be a little over-exaggerated, it nonetheless illustrates a dramatic possibility: much of our health outcomes may be determined by the little guys who call us home.
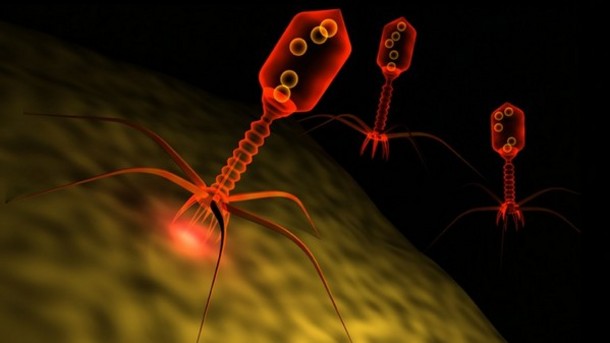
Not only do various organs and cavities of our bodies play host to a panoply of microbiomes, but many of these bacterial act as hosts to phages, which are viruses that target bacteria (you know, the ones that look like the Apollo lander). By examining previously unexplainable sequence data, Edwards et al pieced together the genome sequence of a novel phage. They dubbed it crAssphage - - not because it was discovered in fecal samples ... or because Edwards lab members collected sewage samples across Mexico (although both of these are true) - but because its discovery was enabled by the method of Cross Assembly.
In the last few months, their research has consistently shown that about half of all people carry crAssphage. To take this research global, Rob has pioneered the global crAssphage survey, in which researchers across the globe have started copying Rob’s predilection for politely asking the local sewage company for a wee sample of their finest sludge, and started sending the sequences to him – which incidentally he keeps on a public github account
What they are finding is that just like human genomes across large regions, crAssphage evolves regional differences. While the story keeps getting more and more interesting, the biggest questions have yet to be conclusively answered: what is the host bacterium? What are implications for absence or presence of crAssphage, or variations in its genome? Are their any health risks, or potential medical uses, that could be inferred from this research?
Interestingly enough, a young girl in the audience asked what Rob dubbed the $20 Million dollar question of the night:
What happens when different forms of crAssphage interact within us? We don't know the answer to that question, though Rob's team is working on finding a solution.
Such relationships between and amongst host and phage, and related questions of variability, are already the target of the next step in crAssphage research. That girl’s question capped a night that surely illustrates the bright future of STEM education for our youngsters, and the dynamic community of Microbiome and Bioinformatics scientists in San Diego.