Earlier this month, we launched NanoString Masterclass Series, designed to enhance scientific understanding and practical skills in advanced nCounter data analysis. Dr. Ryan Friese (VP of Rosalind Customer Experience) hosts the series to share his best practices from years of working hands-on with scientists in BioPharma and Academia as a Field Application Scientist for NanoString.
Video Highlights (5 mins) - access the FULL replay below
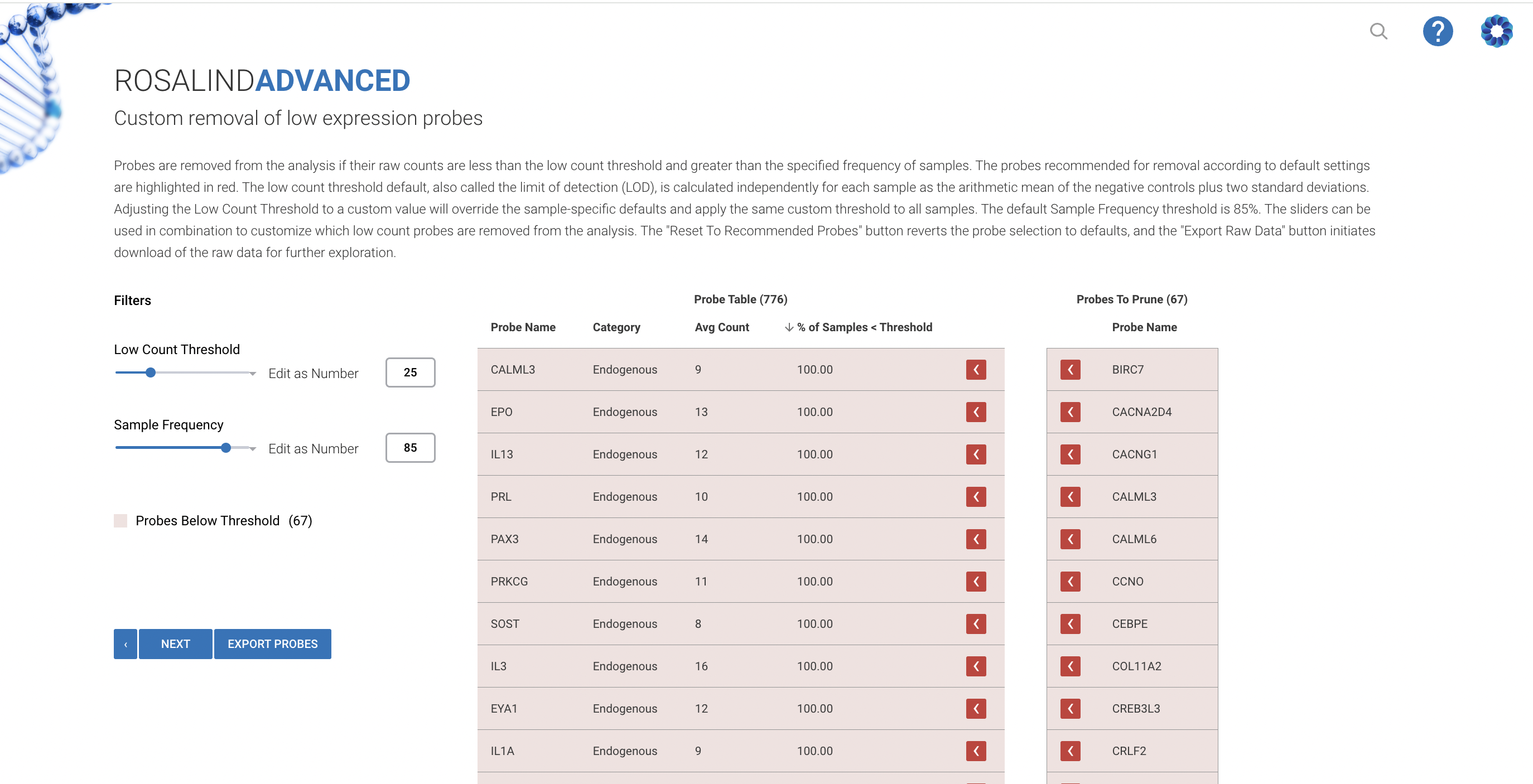
Importance of Custom Pruning
Custom pruning of low expressors is crucial in genomic research, as it helps to significantly reduce background noise, thereby enhancing the overall clarity of the data. This selective removal focuses analysis on genes with higher expression levels, which prevents the potential biases caused by random fluctuations in genes with low expression.
The Rosalind platform simplifies this essential step by employing advanced algorithms designed to intelligently identify and eliminate these low expressors. This automation not only streamlines the research workflow but also substantially improves the quality of the data.
Key benefits of pruning low expressors include:
- Enhanced Data Clarity: By filtering out statistical noise, the genuine signals from highly expressed genes are more discernible, making the data cleaner and clearer.
- Improved Analysis Accuracy: With less clutter in the dataset, analytical algorithms can model gene expression relationships more accurately, leading to more accurate results.
- Increased Computational Efficiency: Reducing the number of data points to process can significantly speed up computational tasks, allowing for faster iterative analysis, which is vital in high-throughput environments.
Algorithms in Custom Pruning (downloadable Resources)
During the webinar, Ryan demystified the complex algorithms and calculations integral to the custom pruning process. For those interested in a deeper dive, we have made the relevant resource files available for download. These materials will allow you to closely follow the methodologies discussed and apply the concepts alongside the webinar replay.
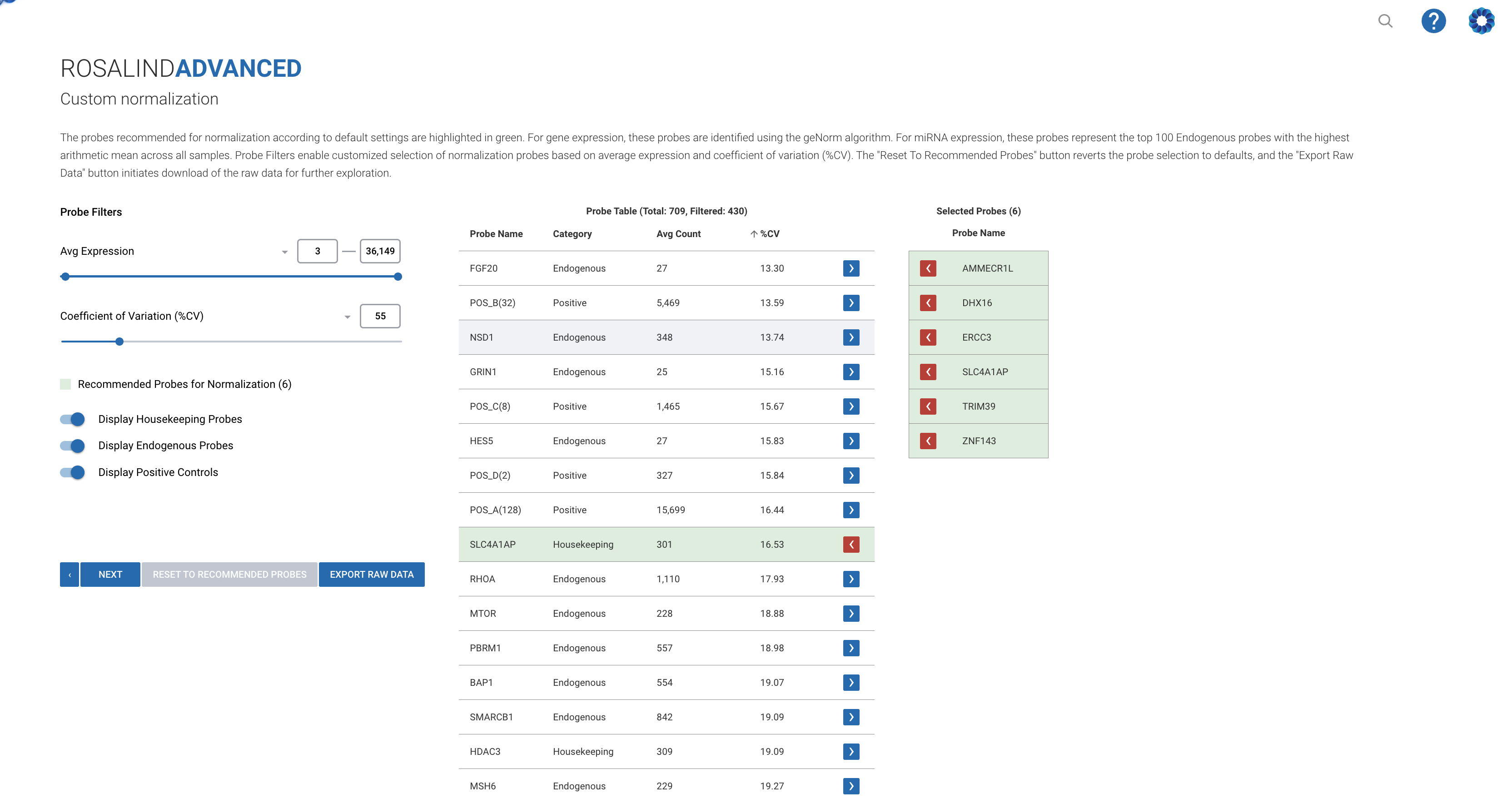
Why Rosalind Enhances Pruning Efficiency
The Rosalind Advanced NanoString Data Analysis module enhances the pruning of low-expression probes by utilizing sophisticated algorithms combined with a user-friendly interface:
-
Intelligent Pruning: Automatically applies best-practice methods and advises on optimal thresholds for pruning, tailored specifically to each dataset and research objective.
-
Visual Feedback and Tools: Offers interactive tools and real-time visual feedback, helping researchers see and understand the impact of pruning on their data analysis immediately.
-
Seamless Integration: The pruning process is fully integrated with Rosalind’s comprehensive suite of tools for analysis, visualization, interpretation, and collaboration. This integration ensures a smooth, efficient workflow from data preprocessing to final analysis, enhancing productivity and reducing the potential for errors.
Benefits for All Levels of Expertise
This masterclass series is designed to benefit all scientists, regardless of experience level. Whether you are deeply experienced in nCounter (nSolver) data analysis or are just beginning to explore this field, the techniques demonstrated can dramatically enhance your research methods and outcomes.
Replay Now Available
Couldn’t attend the live session? No worries! You can watch the replay by clicking the button below. This is a great opportunity to understand in-depth the advanced techniques discussed and learn how they can be applied effectively in your own research projects.
Join the NanoString Masterclass Series
This session was just the beginning! We are excited to continue this series with more in-depth explorations of other aspects of NanoString data analysis. Stay tuned for future sessions where we will cover topics like data integrity, scalability of experiments, and more.