Our mission is to transform how cancer researchers and biologists analyze their data (at SCALE)!
Advanced bioinformaticians deserve all of the credit for building state-of-the-art applications to analyze genomic, proteomic, and microarray data. While these tools remain the lifeline of genomic analysis, more simplified user experiences are now required to empower today’s cancer biologists to focus on their research application, not which software application to use.
Tweet: The self-guided Rosalind™ platform empowers #researchers to analyze their own #genomic data. @OnRampBio
.png?width=768&height=581&name=CheckOutRosalindatOnRampBio%20(1).png)
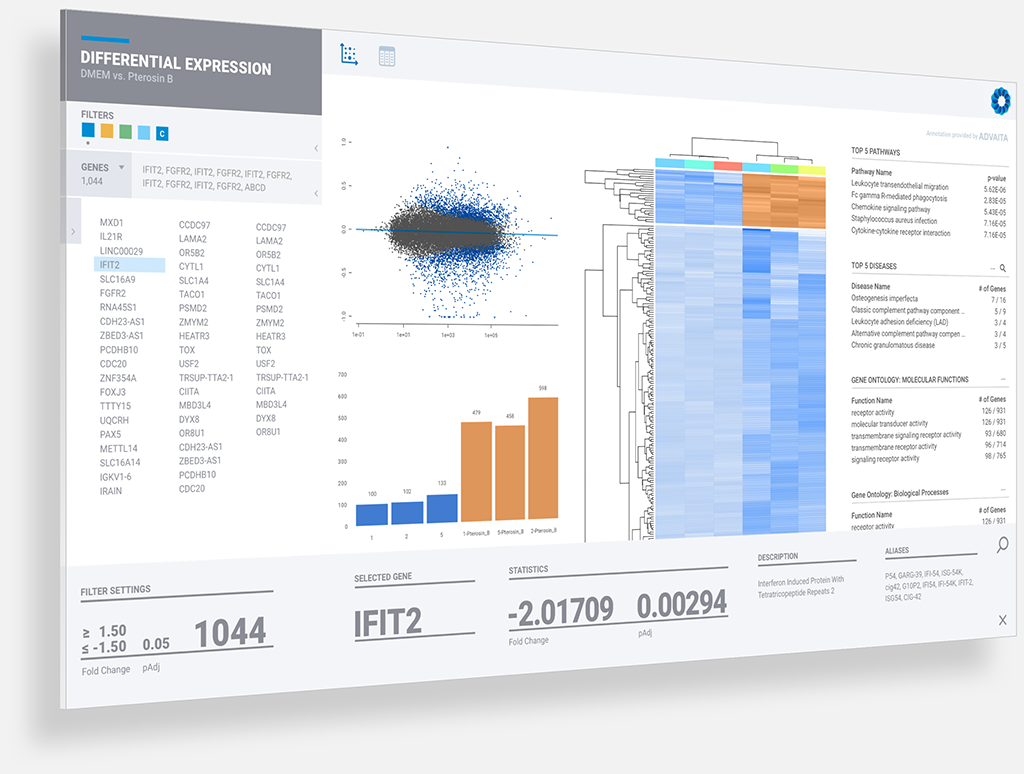
For the first time, researchers performing large-scale genomics studies across a range of fields (including drug development, precision medicine, and agri-business) can rapidly and dynamically explore and interpret their own data, accelerating their next phase of research while saving time and money.
If you are a researcher doing genomic analysis, come see what Rosalind™ can do
(free trial- 4 free samples included!)